#Industry News
As a powerful tool for tumor monitoring, what are the application scenarios of MRD in clinical promotion?
As a powerful tool for tumor monitoring, what are the application scenarios of MRD in clinical promotion?
MRD, from its mature application in the field of hematologic malignancies to its implementation in the clinical practice of solid tumors, is gradually shifting its value from prognosis assessment to predictive potential, spanning across all stages of tumor development. With the initiation of various interventional clinical trials, the precise diagnosis and treatment based on MRD outcomes in the future could potentially reshape the landscape of cancer therapy, greatly benefiting patients' survival. In this issue, we will explore various aspects of MRD through a question-and-answer format to provide comprehensive insights.
Basic knowledge
1. What is MRD?
MRD is described in various ways: Minimal Residual Disease, Measurable Residual Disease, and Molecular Residual Disease. The concept, originally applied to hematologic malignancies, has gradually extended to solid tumors. It primarily refers to cancer-derived molecular abnormalities that cannot be detected by traditional imaging or laboratory methods after treatment but are discovered through liquid biopsies. These abnormalities serve as biomarkers indicating the presence of residual tumor cells in the body following treatment.
2. What is whole exome sequencing (WES), and what is tumor-specific WES?
Whole Exome Sequencing (WES):
a) Sequencing Scope: Encompasses all coding regions of the human exome (over 20,000 genes).
b) Sequencing Depth: Effective average sequencing depth of ≥100×; coverage of at least 95% at 20×.
c) Sequencing Data: Effective data volume of ≥10G.
SpaceGen has independently developed a tumor-specific version of Whole Exome Sequencing (WES) testing. In addition to the standard WES, this approach includes coverage of tumor hotspot mutation regions, intronic regions related to common gene fusions, pathogenic sites in non-coding ClinVar regions, SNP markers, and microsatellite instability (MSI) regions. The results of baseline sample testing cover targeted therapy, immunotherapy, and genetic risk assessment.
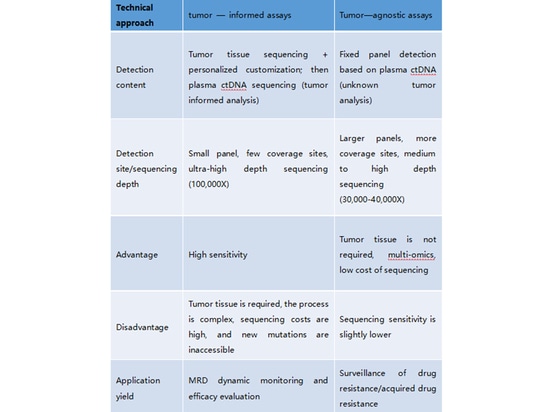
3.What is tumor-informed analysis, and what is tumor-agnostic analysis? What are their characteristics?
Tumor-Informed Analysis (Tumor-Informed Strategy):Tumor-informed analysis, also known as the Tumor-Informed Strategy, involves initially testing a patient's tumor tissue sample to understand specific genomic variations. Subsequently, certain variant sites present in the tumor tissue are monitored using ctDNA (circulating tumor DNA) analysis. In this MRD monitoring approach, the tumor-informed principle is reflected in: (1) the selection and design of ctDNA monitoring sites based on the detected mutations in the tumor tissue, and (2) determining MRD-negative or MRD-positive status based on the detection of tumor-derived mutations in ctDNA. The Signatera product from Natera exemplifies the tumor-informed strategy. It begins with WES testing of the patient's tumor tissue to understand their mutation profile, followed by personalized ctDNA analysis targeting 16 specific variant sites.
Tumor-Naïve Analysis (Tumor-Naïve Strategy):
Tumor-naïve analysis, also known as the Tumor-Naïve Strategy, does not rely on detecting mutations from the patient's tumor tissue. Instead, it involves designing a specialized panel for a specific cancer type and using ctDNA analysis from the patient's plasma. This approach might also incorporate other omics methods, such as epigenetics, alongside ctDNA analysis to determine MRD. Guardant Health's MRD product is a prominent example of the tumor-naïve strategy. In addition to mutation analysis, this product integrates methylation analysis to enhance predictive sensitivity.
4. What are MRD's custom panels and fixed panels?
Custom Panel refers to a panel design that doesn't have fixed genes and loci. It involves performing large panels, whole exome sequencing, or whole genome sequencing on the patient's tissue. Based on factors like the category, type, and abundance of gene mutations, a certain number of personalized or customized genes are selected. These selected positive genes serve as a reference baseline for subsequent regular monitoring and comparison.
Fixed Panel, on the other hand, is designed based on the molecular characteristics of different cancer types. It typically includes driver genes and molecular targets for targeted therapies for various cancer types. A fixed gene/loci panel is used for high-depth sequencing to perform dynamic monitoring.
Both fixed and custom panels can be used in a cross-combination for both Tumor-Informed and Tumor-Naïve strategies. Fixed panels are more commonly applied in cases where tissue samples cannot be obtained, as seen in the Tumor-Naïve strategy.
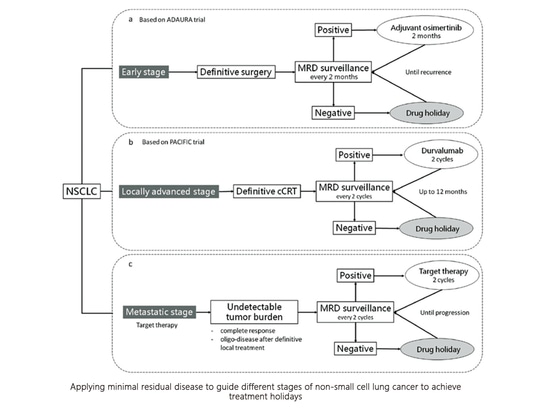
5. What is the drug holiday?
The drug holiday, also known as intermittent treatment, refers to a strategy where a medication is temporarily discontinued after achieving a satisfactory therapeutic effect through long-term usage. Despite the break in medication, the treatment's effectiveness is still maintained. The duration of the drug holiday can range from several days to months and even years. While the concept of drug holidays is not new and has been used in other chronic conditions that require long-term medication, its application in the field of cancer treatment still requires further research.
A small exploratory study conducted at the Guangdong Provincial People's Hospital, reported at WCLC 2021, was part of the CTONG 1602 (NCT03046316) trial.
Criteria for drug holidays include:
1) Absence of visible lesions in imaging (CT, MR, or PET/CT).
2) Negative minimal residual disease (MRD).
3) Negative carcinoembryonic antigen (CEA).
During the drug holiday phase, patients do not receive any treatment but undergo monitoring through CT, CEA, and MRD evaluations every three months.
Criteria for resuming treatment:
1) Disease progression (RECIST PD).
2) Positive MRD.
3) Gradually rising CEA levels.
Clinical application
1. What are the differences between MRD and tumor-targeted sequencing?
Tumor-targeted sequencing primarily focuses on identifying specific genetic mutations in a tumor that are relevant to targeted therapies. It serves as a diagnostic or guiding tool for targeted treatments, and can also be used to study mechanisms of resistance to targeted therapies. Targeted sequencing panels often include other molecular markers such as microsatellite instability (MSI), tumor mutational burden (TMB), and chemotherapy sensitivity assessment.
MRD, on the other hand, aims to detect molecular residual disease. It assesses treatment response, monitors for relapse, and is not specific to a particular treatment modality. Instead, it focuses on comprehensive systemic treatment of malignancies. MRD utilizes personalized or fixed panels of ctDNA variations to dynamically evaluate treatment response and the occurrence of relapse and metastasis for each individual patient.
2. Which cancer types is MRD currently applicable to?
In theory, MRD is applicable to all solid tumors. However, some studies suggest that central nervous system (CNS) tumors might have lower sensitivity. Currently, there have been numerous research results published for cancer types such as lung cancer, colorectal cancer, breast cancer, prostate cancer, esophageal cancer, liver cancer, gastric cancer, and bladder cancer.
3. What are the clinical applications of MRD in its promotion?
Perioperative Comprehensive Management:
a) Assessment of Neoadjuvant Therapy Benefit: Baseline analysis of biopsy samples + dynamic monitoring of neoadjuvant therapy cycles (multiple times).
b) Postoperative Molecular Residual Disease Detection: Postoperative sample baseline analysis + MRD assessment (1 time).
c) Assessment of Adjuvant Therapy (Postoperative/Progression) Benefit: Postoperative/biopsy sample baseline analysis + MRD assessment (multiple times).
d) Relapse Monitoring: Postoperative/adjuvant therapy baseline analysis + MRD assessment (multiple times).
How should the sampling/monitoring time for MRD be selected?
Currently, there are no established guidelines or consensus documents providing standardized recommendations for the timing of baseline sampling and monitoring for MRD. However, the 2021 "Expert Consensus on Molecular Residual Disease in Non-Small Cell Lung Cancer" describes that the half-life of ctDNA is approximately 35 minutes, and the ctDNA status three days after surgery can indicate the risk of relapse (DYNAMIC study). The initial window for MRD testing is typically set within 4 months after curative treatment, as mentioned in the consensus. For locally advanced NSCLC patients receiving curative chemoradiotherapy and preparing for consolidation immunotherapy, the window for MRD testing is defined as at least 1 week after curative chemoradiotherapy (CAPP-Seq study).
Currently, the consensus recommends that the initial MRD testing time window for NSCLC should be within 1 week to 1 month after curative treatment.
4.Can MRD with Tumor-informed strategy be used on patients without tissue samples or treatment?
MRD monitoring using the Tumor-informed strategy requires sequencing of tumor tissue and customizing the MRD detection panel based on the patient's personalized variation information. The limitation of this strategy is tissue dependence, so MRD with Tumor-informed strategy cannot be used without tissue samples.
Treated patients: Tumors have spatial and temporal heterogeneity. Treatment options for different cancer types may change the molecular characteristics of tumors. There are differences between the baseline analysis of previous tumor samples and the molecular characteristics after treatment, which may affect the accuracy of test results. At present, there is no unified opinion in the guidelines and consensus. Recurrent/metastatic lesions can be used for baseline analysis, follow-up dynamic surveillance, or the Tumor-naïve strategy can be used.
6. If the first postoperative minimal residual disease (MRD) is negative, and clinical postoperative adjuvant treatment is administered, is it still necessary to conduct multiple follow-up dynamic monitoring?
A negative MRD result does not mean that the patient is without the risk of recurrence or metastasis. Postoperative negative molecular residual status also does not serve as the sole criterion for assessing the suitability of adjuvant treatment in clinical evaluation. Even if the initial MRD test is negative, it is still advisable to dynamically monitor the recurrence and metastasis situation in subsequent follow-ups, and to proceed with monitoring and diagnosis based on the recommendations of clinical doctors.
SpaceGen
SpaceGen has introduced solid tumor molecular residual disease (MRD) detection services with a wide-ranging product testing panel that supports various solid tumor MRD detections to meet clinical needs.
1. What is the technical approach for solid tumor molecular residual disease (MRD) detection services?
SpaceGen's MRD detection employs a customized panel testing strategy, also known as Tumor-informed assays. It begins by conducting comprehensive whole exome sequencing (WES) on the primary tumor tissue to identify patient-specific genetic mutations. Subsequently, a personalized panel is designed to target a specific number of mutation sites in the tissue for subsequent ctDNA (circulating tumor DNA) testing.
Tumor-focused WES testing (tumor tissue + peripheral blood) → Selection of specific mutation sites → Personalized panel customization → Ongoing multiple monitoring (peripheral blood samples)
2. Project Advantages & Parameters
• Independent development of tumor-focused whole exome sequencing (WES) detection, which extends beyond standard WES by including coverage of tumor hotspot mutation regions, intronic regions relevant to common gene fusions, pathogenic sites in non-coding ClinVar regions, SNP (Single Nucleotide Polymorphism) scaffolds, and MSI (Microsatellite Instability) regions. Baseline sample detection results cover targeted therapy, immune therapy, and genetic risk assessment.
• Personalized customized panels employ in-house developed single primer amplification combined with UMI (Unique Molecular Identifier) sequencing library construction technology.
• Automated monitoring site selection is carried out using weighted scoring based on factors such as mutation frequency, gene function, mutation type, and mutation level. This approach enhances sensitivity in site selection for monitoring.
• Ultra-deep sequencing (100,000X) combined with UMI analysis achieves a mutation detection sensitivity of 0.05%, meeting the requirements for MRD detection performance.
• Baseline analysis plus multiple dynamic monitoring enable comprehensive tumor diagnosis and treatment, offering a complete cycle of MRD dynamic monitoring across the course of cancer treatment.
3. Combination Options
Flexible comprehensive management with multiple choices:
• Whole exome baseline analysis + 1 round of dynamic monitoring
• Whole exome baseline analysis + 3 rounds of dynamic monitoring
• Whole exome baseline analysis + 6 rounds of dynamic monitoring
• Whole exome baseline analysis + N rounds of dynamic monitoring
Applicable for prognosis prediction, recurrence risk assessment, and treatment efficacy monitoring across various solid tumors.